Automatic Cell Image Segmentation Using Genetic Algorithms (source)
Keywords: cell segmentation, genetic algorithms, auto machine learning, auto tuning.
Cell image segmentation is a fundamental stage for cell identification process, but it is not an easy task. Several methods for cell segmentation have been proposed. However, the selection of parameters for the available algorithms depends on the cell type and finally they are designated by an expert. Whereas this approach can result in good performance, it is not necessarily the optimal and may inherit expert’s biases. We propose in this paper an autonomous machine learning technique for the selection of parameters in the cell image segmentation process.
Proposed approach:
In this study we are interested in parameter auto-tuning, more specifically, in a setting where the parameter space of a cell segmentation algorithm is automatically explored by a genetic algorithm. The auto-tuning process compares the results with ground truth, and the process is repeated to find a set of parameters that produces the most accurate results as measured by a comparison metric, that in our case is the F-index.
Cell segmentation algorithm
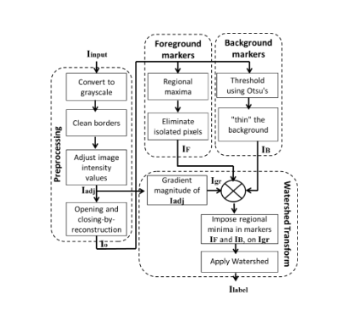
The proposed method to distinguish between background and cell is the marker-controlled watershed (MC-Watershed). The goal of this block is to recognize as many cells as possible. A flowchart of this process is shown in Figure below. Four stages integrate the MC-Watershed: preprocessing, marking the foreground objects, computing background markers and computing Watershed transform.
Fitness function
The fitness function is based on the cell segmentation algorithm. The input vector contains the three parameters r, p and DT, which correspond to radio of the disk, number limit of the pixels to remove elements and option for the distance transform, respectively. The operations described in the flowchart in Figured above are executed. The output image Ilabel is compared with the ground truth and the F-index is obtained. This process is completed for each of the training images (10 images). Then, the average F-index is calculated: it is the output of the fitness function and the value to optimize by the GA
Genetic Algorithm
The GA maps each parameter of our cell segmentation
algorithm to a gene of an individual. This is the input vector to
the fitness function, which perform the segmentation algorithm
and obtain the F-index for each individual. The initial
population is created randomly and evolved using crossover
and mutation. The crossover uses a one-point crossover
between pairs of individuals with a probability of C. The
mutation in each gene of occurs with an independent
probability of M.
In the next figure, the auto-tuning process of the GA is showed. The process takes initial parameters (r, p and DT) as a constraint. The parameters are then introduced into the MC-Watershed algorithm for the cell segmentation process. Afterwards, the Ilabel (segmented image) and Ibinarized (ground truth) are used
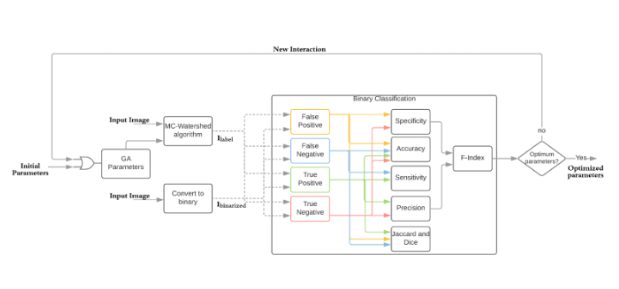
In the block Binary classification, we calculate the specificity, accuracy, sensitivity, precision, Jaccard index, Dice as well of the F-index. The metric to optimize is F-index. Based on the result of the objective function value and the configuration, the GA looks for other individuals in the population, using mutation, crossover, and other operations. This process is repeated until the GA finds the parameters that produces the most accurate results for the F-index.
Results
This section evaluates the auto-tuning algorithm using GA with the goal of maximizing the F-index metric. These experiments were executed with the SNP HEp-2 cell dataset. The dataset (SNPHEp-2) was obtained between January and February 2012 at Sullivan Nicolaides Pathology laboratory, Australia. This dataset contains images of five cell classes: centromere, coarse speckled, fine speckled, homogeneous, and nucleolar; and consists of 1,884 cell images extracted from 40 specimen images. DAPI image channel was used to obtain the cell image masks automatically. With the aim of validating this proposal, 40 cell images from the homogeneous class were randomly chosen.
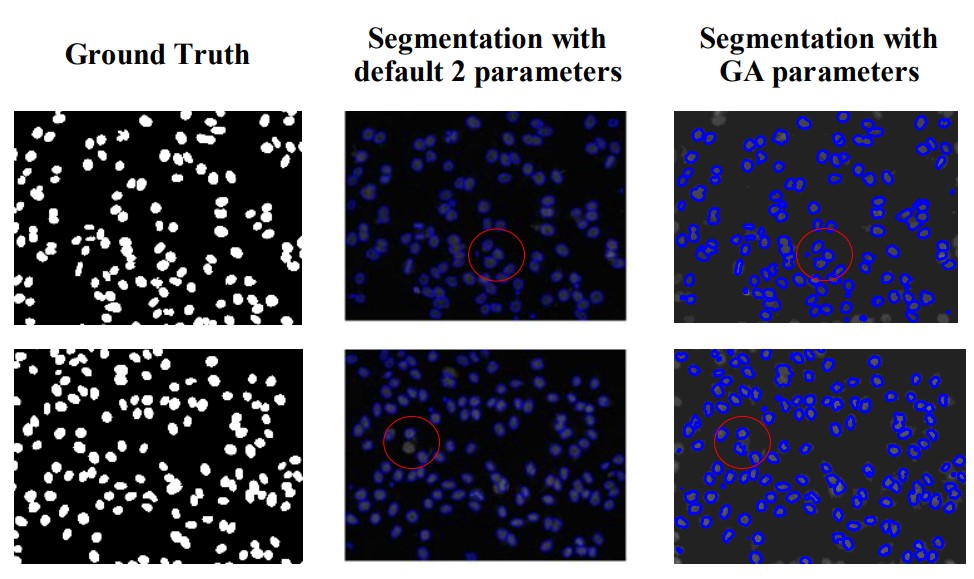
A panel with two images (000002_p2.tif and 00005_p1.tif) is presented in Fig. 3 to show the improvement in segmentation output generated by the tuned versus default input parameters. This image shows that the segmented image with GA reach superior results regard to identification and correct separation of clustered cells. However, some objects were over-segmented.

The performance of the GA was measured with an evaluation of binary class according to the final parameters. The images from the dataset were used for the evaluation. The following metrics were measured: specificity, accuracy, sensitivity, precision, dice and Jaccard index; these, in function of the True Positive (TP), True Negative (TN), False Positive (FP) and False Negative (FN) values. The average values of the metrics for the complete set of images are presented in The next table. The results show that the auto-tuning algorithms improved the quality of the results compared with the results generated by the default input parameters selected by the human expert, regard to F-Index and Jaccard indicator
| Indicator | Method 1 | Method 2 | Method GA |
|---|---|---|---|
| Precision |
0,868049033 |
0,89756878 |
0,879213594 |
| Sensitivity |
0,856911404 |
0,837711863 |
0,853728919 |
| Specificity |
0,973841827 |
0,980786459 |
0,976242719 |
| Accuracy |
0,970271024 |
0,955947701 |
0,954925408 |
| F-Index |
0,953385145 |
0,863037902 |
0,863092628 |
| Jaccard |
0,755985697 |
0,762761452 |
0,762892065 |
| Dicce |
0,858631642 |
0,377202695 |
0,377052532 |
Conclusions
Cell image segmentation algorithms are sensitive to input parameters and the selection done by human experts not always is the optimal option. The proposed auto-tuning algorithm using GA improved the indicators for cell segmentation and it produced accurate visual results. Although it may take computational time, this search method makes it easier for the programmer to select the multiple parameters of the algorithm. As a future work, we intend to apply AutoML for the full selection model problem, which not only include the tuning